ArrayPipe Online Documentation
[← previous] [up ↑] [next →]
Data Input
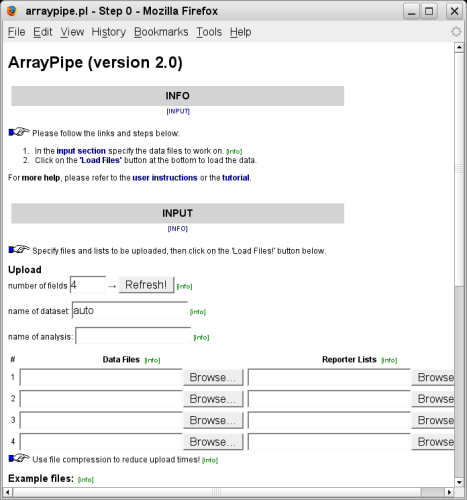
The initial analysis page presents the data loading screen, which looks similar to this:The Info section provides you with information and links that help you navigate to the other sections within the page. Please read it first!
You can upload your own data files that are stored on your local computer or
use the example files that are provided.
Currently, ArrayPipe supports the following file formats:
- ArrayVision
- Imagene (two files per slide!)
- GenePix
- ScanAlzye
- Spotfinder
- Agilent
| Spot ID | CH1I | CH1B | CH2I | CH2B | Flag |
|---|---|---|---|---|---|
| Gene A | 188 | 56 | 276 | 45 | 0 |
| Gene B | 0 | 77 | 9021 | 71 | 1 |
| Gene C | 3020 | 112 | 985 | 98 | 0 |
| etc. | |||||
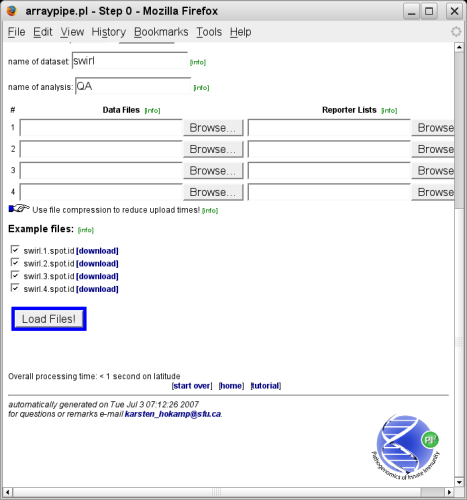
Once you have specified your data input, click on the button labeled 'Load Files!' near the bottom of the page (a couple of example files might be listed as well):
Specifying names for the dataset and the analysis is useful if your ArrayPipe version has a database back-end in place.
Tip:To save time and bandwidth you can upload compressed data files. You can also combine multiple files into a compressed archive using tools, such as 7-zip or tar.
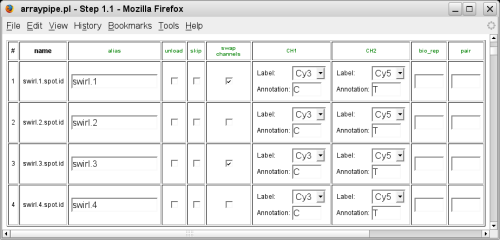
Once the data are loaded, you can re-name the slides, select dye-swaps, annotate your channels and specify biological and technical replicates:
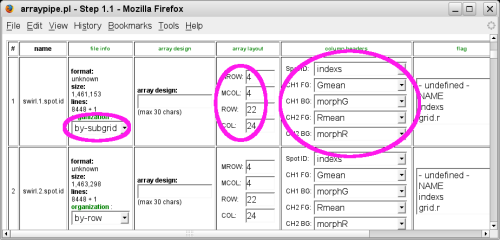
It is also very important to get the array dimensions and file organization right:
- Some quantification programs store the spot entries by subgrid, some store them by row (across the whole slide).
- The grid layout (metarow, metacolumn, row and column) is important for functions that work grid-specifically, e.g. printTipLoess normalization.
- Make sure that the column headers are set so that you read in the correct data.
Note:
The above settings can be stored in ArrayPipe so that you don't have to
manually specify them everytime you load data files from a specific
array. Please speak to the person administrating your version of ArrayPipe to
arrange for that.
[← previous]
[up ↑]
[next →]
Home
 last modified $Date: 2007/08/27 14:02:03 $
last modified $Date: 2007/08/27 14:02:03 $
for questions or remarks e-mail karsten_hokamp@sfu.ca.